Research Overview
The Luna Lab investigates how RNA-binding proteins (RBPs) influence viral infections and cellular stress responses at the subcellular level. Using positive-sense RNA viruses as models, we explore how RBPs orchestrate viral replication and and modulate innate immune responses. Through innovative techniques in reverse genetics, CRISPR screening, and subcellular RNA profiling, we map the dynamic spatial interactions between viral genomes and host RBPs throughout the infection cycle. Our goal is to uncover the regulatory logic governing these interactions within distinct cellular compartments, providing insights into fundamental RNA biology and potential therapeutic targets for viral infections and RBP-related disorders.
In addition, we are committed to training and mentoring the next generation of scientists in biomedical research, particularly from under-represented groups. We further prioritize scientific outreach efforts at all levels to educate, inform and inspire.
What we’re working on
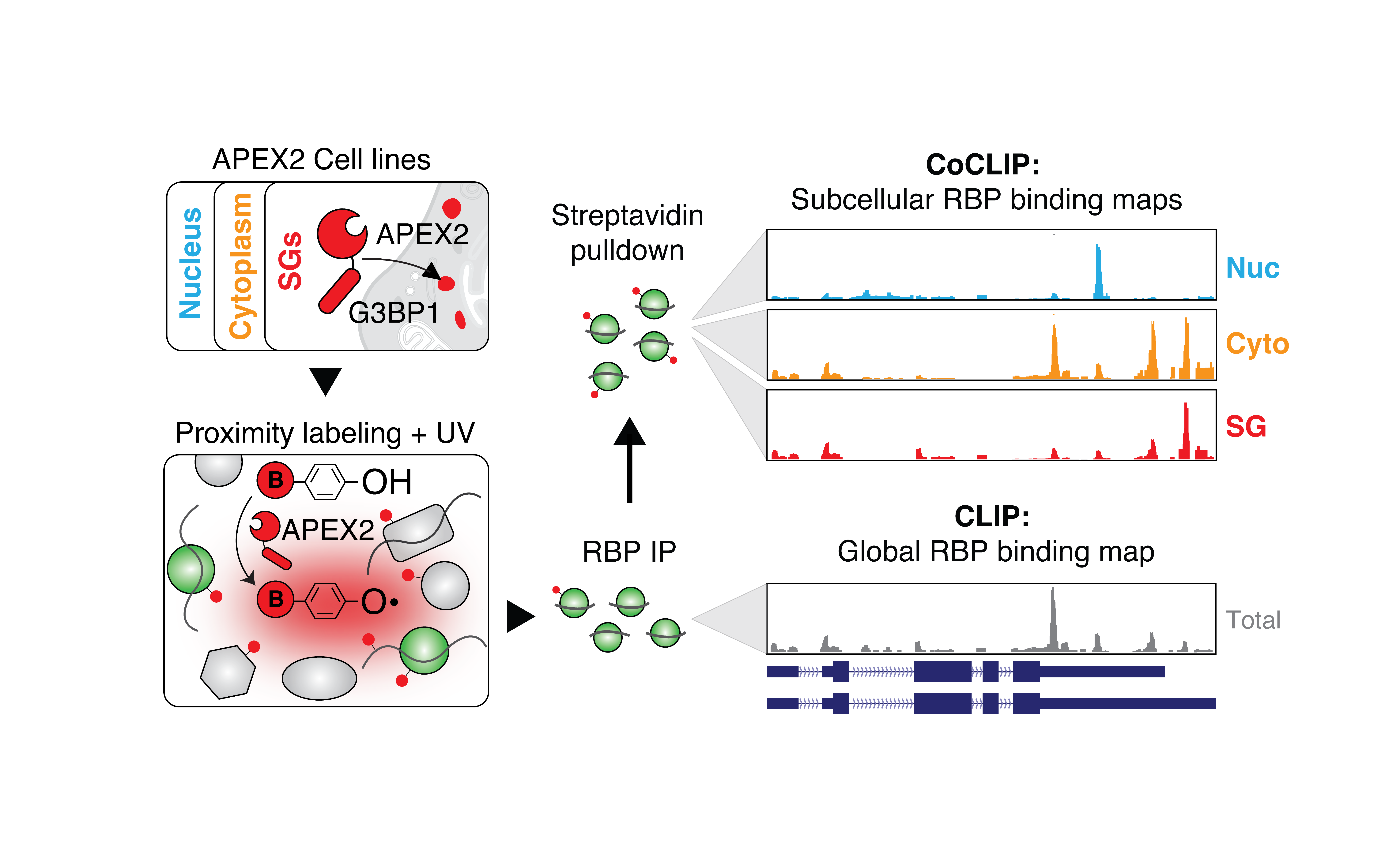
Mapping Subcellular RBP:RNA Interactions: We’re developing innovative tools to capture and analyze RBP:RNA interactions with unprecedented subcellular resolution. By biochemically preserving spatial information in living cells, we aim to uncover how the localization of these interactions influences cellular functions and viral infections. This project will provide crucial insights into how RBPs operate in distinct cellular compartments and how their spatial distribution affects their roles in RNA metabolism.
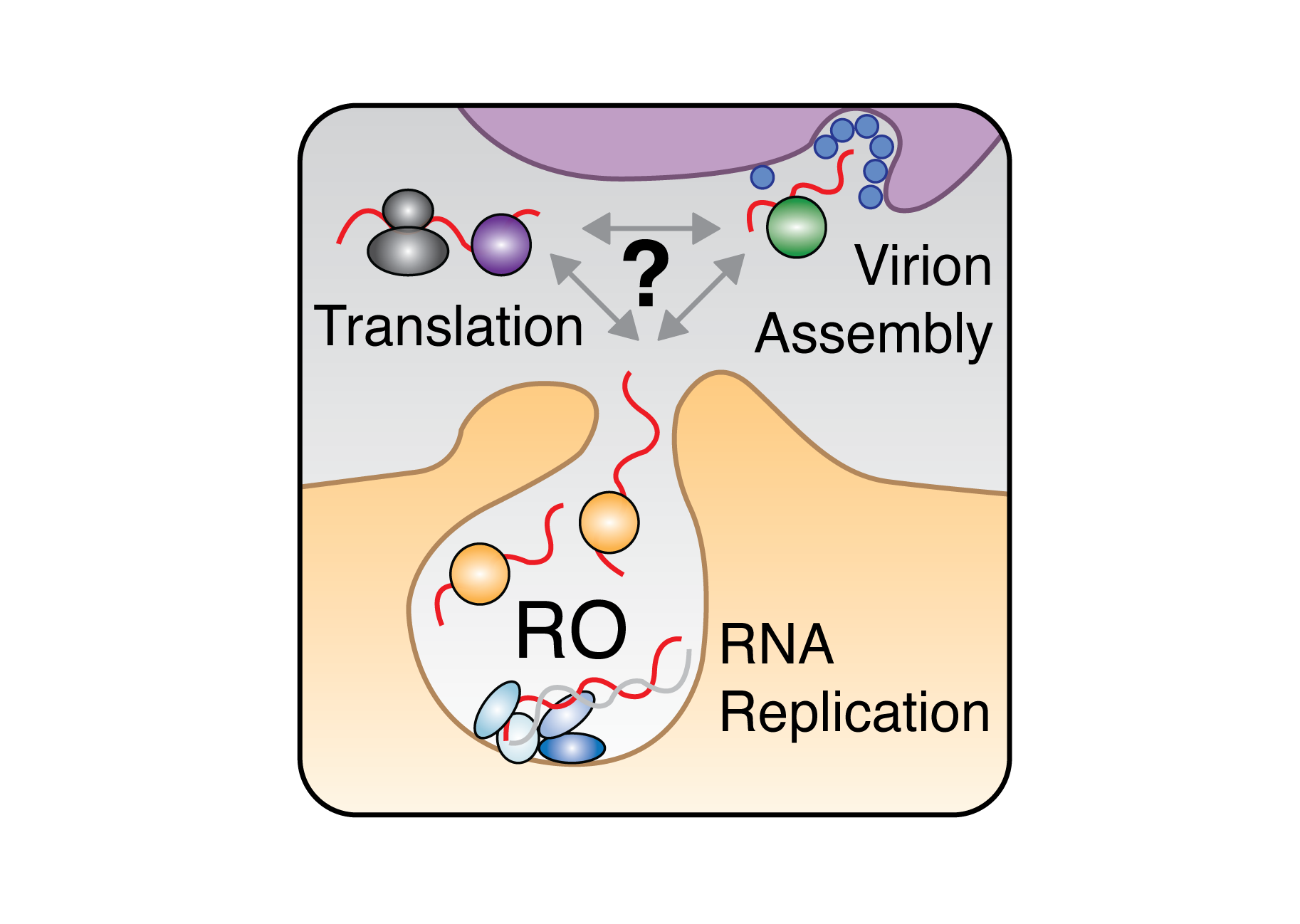
Decoding RBP Dynamics in Viral Infections:
Using positive-sense RNA viruses as model systems, we’re investigating how RBPs how cellular RBPs orchestrate the balance between viral genome replication, translation, and virion assembly. We use viral reverse genetics, CRISPR screening, and advanced RNA profiling to map the dynamic interactions between viral genomes and host RBPs throughout the infection cycle. Our goal is to unravel the regulatory logic of RBPs during infection with an eye toward novel antivirals and therapeutics.
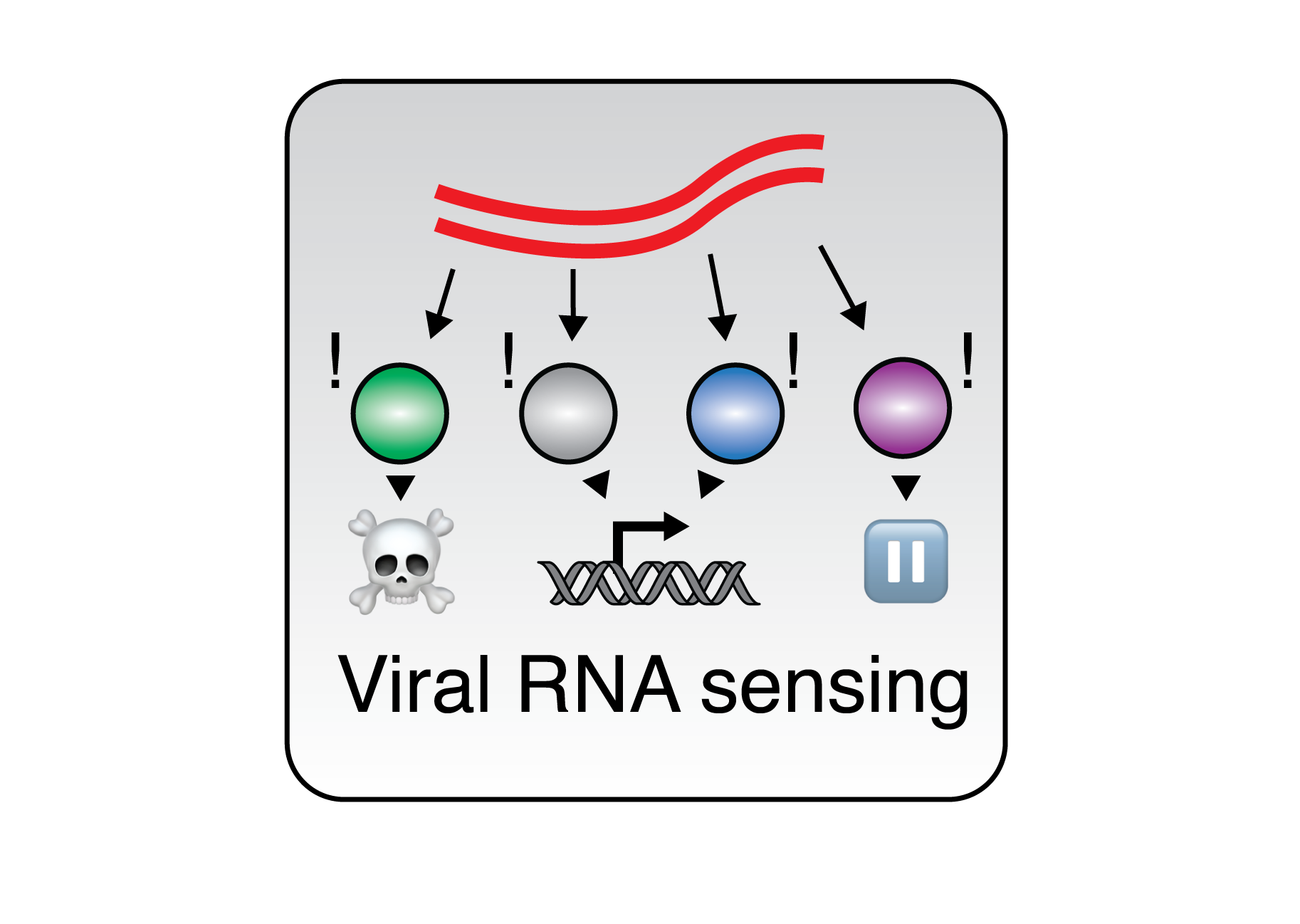
Modeling RBP Networks during Innate Immunity Host recognition of viral RNA by RBPs is among the foundational steps of innate immune sensing, yet how this process is coordinated among RBPs with different downstream outcomes is unknown. We use our experimental data with state-of-the-art bioinformatics tool to build models of RBP activities and interaction networks during changing cell states. By analyzing how RBP:RNA interactions change in response to infection, we aim to predict key regulatory nodes and potential vulnerabilities in the host-pathogen interface, guiding future experimental validations and therapeutic strategies. This work bridges our experimental work with a systems-level understanding of RNA regulation.